Abstract
Paxillin, a member of the group 3 subfamily of LIM domain proteins, is localized within focal adhesions and participates in a number of signal transduction pathways mobilized upon activation of cell surface receptors. In recent years, a number of group 3 LIM domain proteins have been found to also localize within the nucleus and exert direct effects on transcription. We show here that paxillin is present within nuclei and can target the nuclear matrix of CV-1 cells, cultured prostate cancer cell lines, and human prostate tissue. The increased targeting of androgen receptor to the nuclear matrix upon overexpression of paxillin may be brought about by direct interactions between paxillin and the receptor, which were detected in vitro. Paxillin functions as a coactivator for androgen receptor and glucocorticoid receptor, but not estrogen receptor α, similar to its close relative Hic-5/ARA55. Both paxillin and Hic-5/ARA55 use their COOH-terminal LIM domain to interact with steroid receptors. However, paxillin is distinguished from Hic-5/ARA55 by both the location of its receptor coactivation domain (i.e., COOH-terminal LIM domain) and by the dominant-negative activity of its NH2-terminal domain. Thus, highly related group 3 LIM domain proteins may use distinct mechanisms to modulate steroid hormone receptor transactivation.
INTRODUCTION
Androgens are required for the growth and development of the normal prostate gland (1) but also participate in the progression of prostate cancer, particularly in early stages (2). The principal mediator of androgen action within cells is the AR,3 a member of a large superfamily of nuclear receptors (3, 4). Physiological effects of androgens result primarily from transcriptional regulation of specific target genes brought about by AR interaction with specific DNA sequences or receptor associations with other transcription factors (5, 6, 7).
The NH2- and COOH-terminal regions of nuclear receptors, including ARs (8, 9, 10, 11), possess transactivation domains that function to either enhance the association of basal transcription factors with the preinitiation complex (12) or recruit various coactivator or coactivator complexes (13). Many coactivators exert their actions through the modification of histones (14, 15) or transcription factors (16, 17, 18). Families of nuclear receptor coactivator proteins have been identified that either directly or indirectly affect protein acetylation (13) or methylation (19, 20). Although the modulation of chromatin structure at hormonally responsive promoters is a critical feature of nuclear receptor coactivators, it seems likely that distinct coactivators could use different mechanisms to potentiate nuclear receptor transactivation (10, 21, 22).
The AR has proven to be a particularly useful reagent for the identification of novel transcriptional coactivators (23, 24, 25, 26). For example, Hic-5/ARA55 was first identified as a coactivator for AR (27) and later shown to potentiate the transactivation activities of a subset of steroid hormone receptors (28) including the GR. Although Hic-5/ARA55 was shown to associate with the nuclear matrix (28), most studies of this protein have focused on its focal adhesion binding (29, 30). Hic-5/ARA55 is a member of the paxillin family of focal adhesion proteins (31, 32) that share a closely related COOH-terminal LIM domain region (33). The LIM domain is a unique double zinc finger motif (33) with a conserved CXXCX-16–23 amino acid–HXXHXXHXXHX-16–21 amino acid–CXX(D/H/C) sequence and is found in a variety of proteins of diverse functions and localization (34). Paxillin and Hic-5/ARA55 belong to a group 3 subfamily of LIM domain proteins and contain either three or four separate LIM domains in their COOH-terminal region (35).
In addition to their localization within focal adhesions, members of the group 3 LIM domain family are also associated with the nucleus (30, 36). For zyxin and the closely related LPP protein, nuclear export signal sequences have been identified that regulate their shuttling between the nucleus and focal adhesions (36, 37). In addition, transactivation domains have been identified within LPP and another zyxin family member, Trip6 (38, 39). Trip6 was isolated based upon its hormone-dependent interaction with thyroid hormone receptor (40). Finally, although the transcriptional regulatory properties of the zyxin family member Ajuba have not been revealed, nuclear localization of Ajuba LIM domains is required for PC19 embryonal carcinoma cells to undergo differentiation in vitro into an endodermal cell type (41). Despite the prevalence of nuclear group 3 LIM domain proteins, the physiologically relevant transcriptional targets regulated by these proteins remain largely undefined.
In this report, we examine the nuclear localization and steroid receptor coactivation activity of paxillin. Paxillin, similar to Hic-5/ARA55, is a selective steroid receptor coactivator that has the capacity to localize to the nuclear matrix in prostate cancer cell lines and prostate tissue. However, paxillin is distinguished from Hic-5/ARA55 in the domain requirements of its nuclear functions. Thus, highly related group 3 LIM domain proteins may use distinct mechanisms to modulate steroid hormone receptor transactivation. Given the alterations in focal adhesion signaling and paxillin activity in prostate cancer cells (42, 43), the ability of paxillin to function as an AR coactivator in prostate cancer cells could have important implications for the progression of AR-dependent tumors.
MATERIALS AND METHODS
Plasmids.
Mammalian expression vectors for FL paxillin, its NH2 terminus, and COOH terminus with an NH2-terminal HA epitope tag were constructed by inserting PCR-amplified cDNA fragments into the pSG5.HA expression plasmid (28). The NH2-terminal paxillin expression plasmid includes human paxillin amino acids 1–307, whereas the COOH-terminal expression plasmid includes amino acids 308–557. Mammalian expression vectors for FL Hic-5/ARA55 and GRIP1 containing an HA epitope tag were provided by Dr. M. R. Stallcup (University of Southern California School of Medicine, Los Angeles, CA). pLC-LUC contains a luciferase reporter gene driven by the MMTV long terminal repeat with a bifunctional androgen response element and glucocorticoid response element (28). ERE-LUC contains an estrogen response element-driven luciferase reporter gene and was provided by Dr. M. Nichols (University of Pittsburgh School of Medicine). hCMV5-AR contains human FL human AR cDNA under the control of CMV promoter and was provided by Dr. D. Robins (University of Michigan Medical School, Ann Arbor, MI). 6R-GR contains cDNA encoding FL rat GR under the control of Rous sarcoma virus long terminal repeat promoter and was provided by Dr. K. Yamamoto (University of California School of Medicine, San Francisco, CA). CMV-ER containing human ER cDNA under the control of CMV promoter was provided by Dr. M. Nichols (University of Pittsburgh School of Medicine, Pittsburgh, PA). The vectors for FL paxillin, pGEX-4T FL paxillin, pGEX-4T NH2-terminal paxillin, and pGEX-4T COOH-terminal paxillin were provided by Dr. S. Thomas (Harvard Medical School, Boston, MA).
Transient Transfection.
Cells (1 × 105) were passed onto 30-mm cell culture dishes 24 h before transfection. Transient transfections were performed using the Lipofectamine Reagent (Life Technologies, Inc., Gaithersburg, MD). Briefly, cells were incubated in transfection medium for 5 h. The transfection medium was replaced with 5% charcoal-stripped FBS in the absence or presence of 100 nm of DHT, 1 μm Dex, or 10 nm 11β-estradiol. Cells were harvested after an additional 24-h incubation at 37°C in humidified air containing 5% CO2, and then extracts were prepared for luciferase assays (28) using procedures specified by the manufacturer of the Luciferase Assay kit (Promega Corp., Madison, WI).
Nuclear Matrix Preparation.
Subcellular fractionations were performed using procedures described previously (44). Briefly, harvested cells were treated with ice-cold CK buffer (10 mm PIPES, pH 6.8, 100 mm NaCl, 300 mm sucrose, 3 mm MgCl2, 1 mm EGTA, 0.5% and Triton X-100) containing 5 μg/ml aprotinin, leupeptin, and pepstatin, 1.2 mm PMSF, and 4 mm vanadyl riboside complex. After centrifugation, supernatants were saved (i.e., CSK-extract), and pellets were treated with DNase I digestion buffer (10 mm PIPES, pH 6.8, 50 mm NaCl, 300 mm sucrose, 3 mm MgCl2, 1 mm EGTA, and 0.5% Triton X-100) containing 300 units/ml DNase I, 5 μg/ml aprotinin, leupeptin, and pepstatin, 1.2 mm PMSF, and 4 mm vanadyl riboside complex. Resultant pellets were further extracted with 0.25 m ammonium sulfate, and the final pellet after centrifugation was designated as the nuclear matrix fraction. The CK buffer extracts, DNase I digestion extracts, and nuclear matrix proteins were subjected to Western blot analysis using a mouse anti-paxillin antibody (Transduction Laboratories, Lexington, KY), goat anti-lamin B antibody (Santa Cruz Biotechnology, Inc., Santa Cruz, CA), or mouse anti-actin antibody (Santa Cruz Biotechnology, Inc.). For in situ extractions, analogous extractions were performed with cells that were grown on coverslips. An analogous method (45) was used to prepare nuclear matrix fractions from human prostate tissue obtained from radical prostatectomy samples and prostates obtained from organ donors. All tissues were collected after appropriate consent under an Institutional Review Board-approved protocol.
GST Pull-Down Assay.
CV-1 cells were transfected with the pCMV5-AR plasmid and incubated in DMEM containing 10% FBS for 48 h after transfection. CV-1 cells and GRH2 cells were serum starved overnight, followed by an additional 2-h incubation with 100 nm DHT or 1 μm Dex, respectively. Whole-cell lysates were then prepared with lysis buffer (10 mm HEPES, pH 7.0, 450 mm NaCl, 5 mm EDTA, 0.05% SDS, 1% Triton X-100 for CV-1 cells; 50 mm Tris-HCl, pH 7.5, 450 mm NaCl, 2.5% sodium deoxycholate, and 1% NP-40 for GRH2 cells) containing 10 μg/ml aprotinin, leupeptin, and pepstatin, 1 mm PMSF and sodium orthovanadate, and precleaned with glutathione-Sepharose. Twenty-five μl of glutathione-Sepharose was incubated with 125 μg of GST-FL paxillin, GST-NH2-terminal paxillin, GST-COOH-terminal paxillin, or GST protein for 1 h at 4°C. Glutathione-Sepharose-bound GST proteins were washed three times with PBS and then added to the CV-1 or GRH2 cell lysates containing 500 μg of total protein. Mixtures were incubated for 3 h at 4°C and then washed three times with washing buffer. Proteins bound to GST-protein derived from AR-transfected CV-1 cells or GRH2 cells were subjected to Western blot analysis using the AR (46) anti-AR antibody (Santa Cruz Biotechnology, Inc.) or BuGR2 anti-GR antibody (Affinity Bioreagents, Golden, CO), respectively.
IIF.
IIF assays were carried out as described previously (28). Briefly, CV-1 cells were fixed with 4% paraformaldehyde and then permeabilized with 0.1% Triton X-100. An anti-paxillin mouse monoclonal antibody (Transduction Laboratories) or an anti-Hic-5/ARA55 rabbit polyclonal antibody (kindly provided by Dr. K. Tachibana, Harvard University, Cambridge, MA) was used to detect endogenous paxillin or Hic-5/ARA55, respectively, for in situ assays. A FITC-conjugated goat-anti-mouse IgG F(ab′)2 antibody (Chemicon International, Temecula, CA) or a FITC-conjugated goat-anti-rabbit IgG (H+L) antibody (Jackson ImmunoResearch Laboratories, West Grove, PA) was used as a secondary antibody.
RESULTS
Localization of Paxillin within the Nuclear Matrix.
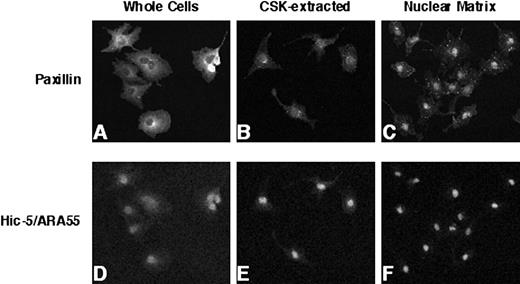
Given the increasing evidence for nuclear localization of group 3 LIM domain proteins (28, 36, 37, 39), we evaluated whether paxillin was likewise present within the nucleus. Biochemical extractions performed either in situ or with harvested cells were used to provide a more thorough assessment of subnuclear compartmentalization of paxillin. As shown in Fig. 1, very little paxillin could be detected within the nucleus of CV-1 cells using IIF analysis with paraformaldehyde-fixed and Triton X-100 permeabilized cells. However, paxillin nuclear staining was revealed when cells were extracted with a low-salt detergent buffer (Fig. 1,B, CSK-extracted). Furthermore, when CV-1 cells were subjected to more extensive biochemical extractions in situ, robust staining of paxillin was detected in the nuclear matrix fraction (Fig. 1,C). The staining pattern of Hic-5/ARA55 in CV-1 was analogous to that observed for paxillin, although nuclear localization of Hic-5/ARA55 was more obvious in whole cells (Fig. 1 D).
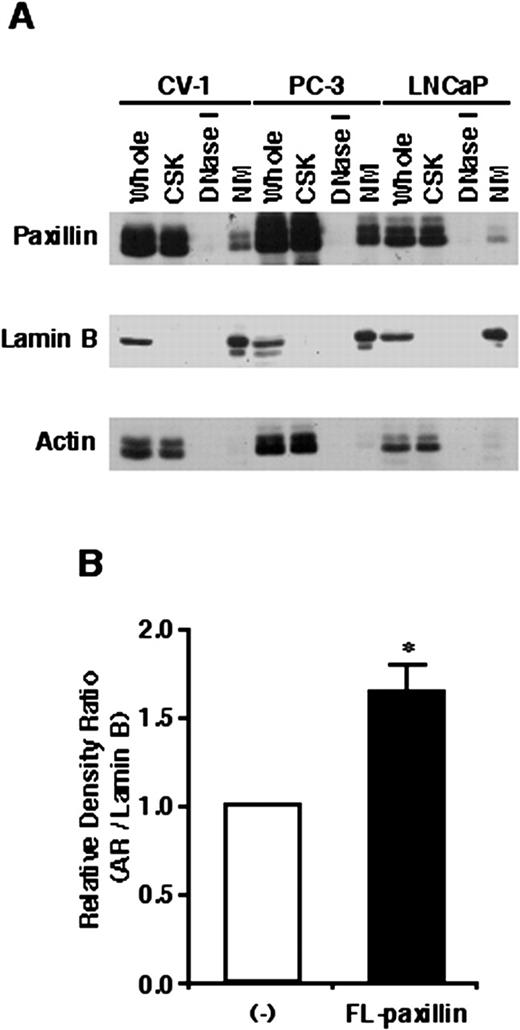
To confirm the IIF results of paxillin localization and broaden the examination of its subnuclear targeting, CV-1 cells and the PC-3 and LNCaP prostate cancer cell lines were harvested and subjected to differential biochemical extractions that ultimately resulted in the isolation of a nuclear matrix fraction. As shown in Fig. 2,A, most endogenous paxillin in all three cell lines examined was recovered from cells by the CSK buffer extraction. Very little paxillin that resists this extraction remains associated with chromatin because DNase I digestion did not lead to recovery of additional paxillin (Fig. 2,A). However, paxillin is detected in the nuclear matrix fraction that remains after extensive biochemical extraction in the three cell lines (Fig. 2,A). The amount of paxillin recovered in the nuclear matrix fraction is estimated from densitometric analysis of these blots to be ∼10% of whole-cell paxillin. This agrees well with previous estimates of the fraction of endogenous Hic-5/ARA55 that targets the matrix of fibroblast cells (28). The integrity of the various biochemical fractions was confirmed upon costaining of blots with either lamin B, to detect nuclear matrix, or actin, to detect CSK buffer-extractable protein (Fig. 2,A). Because nuclear matrix binding of ARs is well established (47, 48), we examined whether paxillin could function as a nuclear matrix acceptor (47) for AR. As shown in Fig. 2 B, overexpression of paxillin in CV-1 cells leads to an increased association of AR with the nuclear matrix. Although paxillin may not be the sole nuclear matrix acceptor protein for AR, these results suggest that it may play a role in the recruitment of AR to the nuclear matrix.
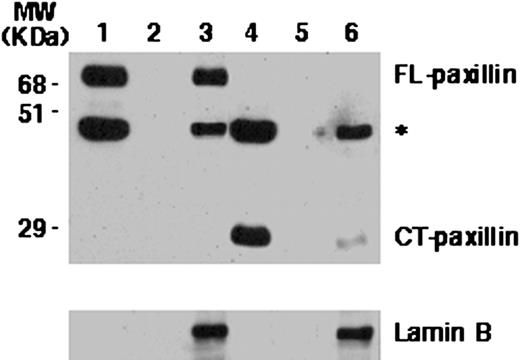
The NMTS of Hic-5/ARA55 is contained within its COOH-terminal LIM domain region (28) and appears to be restricted primarily to its LIM3 domain.4 The NH2-terminal regions of paxillin and Hic-5/ARA55, devoid of LIM domains, are localized exclusively within the cytoplasm when expressed as green fluorescent protein chimera (30). Given the high degree of relatedness of Hic-5/ARA55 and paxillin LIM domains (30), we expected that the paxillin NMTS would likewise be located within its COOH-terminal LIM domain region. However, unlike the COOH-terminal LIM domain region of Hic-5/ARA55 expressed in CV-1 cells, which is indistinguishable from FL Hic-5/ARA55 in its nuclear matrix targeting (28), the COOH-terminal LIM domain region of paxillin is only minimally associated with the nuclear matrix (Fig. 3). Thus, although the NH2-terminal region of paxillin does not appear to target on its own to the nucleus, it must contain sequences that cooperate with the LIM domain region to generate a fully functional NMTS.
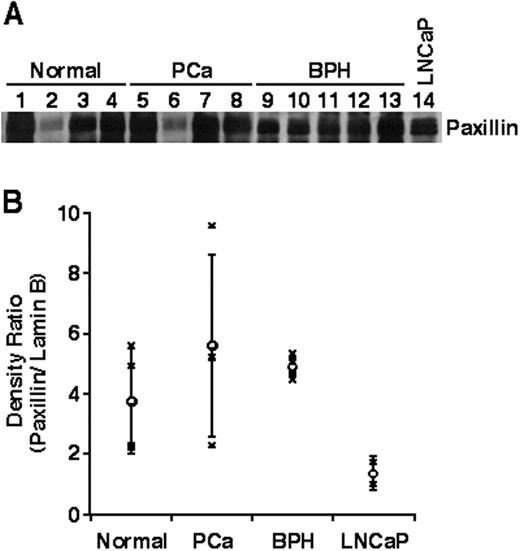
In addition to examining subnuclear compartmentalization of paxillin in prostate cancer cell lines, we performed an analysis of paxillin localization in prostate tissue from normal, prostate cancer, and benign prostate hyperplasia tissue. As shown in Fig. 4,A, paxillin could be detected in the nuclear matrix fraction prepared from prostate, irrespective of the disease state of the tissue. Although our sample size is too limited to make any firm conclusions regarding the relative amount of paxillin in the nuclear matrix of the tissue samples, in just about every case the relative amount of paxillin in the nuclear matrix fractions, expressed as a ratio of paxillin:lamin B, exceeds that detected in LNCaP cells (Fig. 4 B). It is unclear whether apparent differences in the range of paxillin nuclear matrix binding between the various tissue samples are significant. Although the analysis of additional prostate tissue samples will be required to reveal whether quantitative differences in nuclear matrix binding exist in different stages of prostate cancer, our studies clearly establish a nuclear matrix association of paxillin in human prostate tissue.
Paxillin Is a Coactivator for AR.
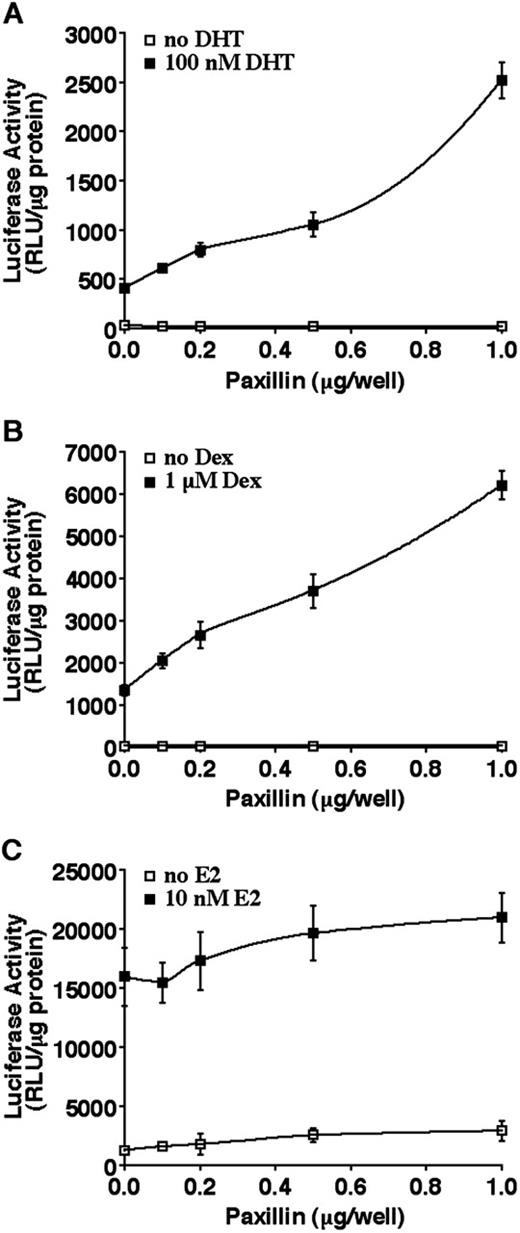
Although Hic-5/ARA55 and paxillin are closely related in their LIM domain structure (30), they exhibit opposing physiological effects, particularly with regard to their role in cellular proliferation and signal transduction (29, 49, 50). It is assumed that much of this difference relates to their divergent NH2-terminal domains (30). Given the ability of paxillin, similar to Hic-5/ARA55 (28), to localize within the nuclear matrix, we examined whether paxillin shared steroid receptor coactivator activity with Hic-5/ARA55. Because paxillin is ubiquitously expressed (51), our studies tested whether overexpression of paxillin was capable of enhancing AR coactivation. As shown in Fig. 5,A, the transactivation activity of AR on the androgen-responsive MMTV promoter was potentiated in transiently transfected CV-1 cells upon cotransfection with a paxillin expression vector. A maximum 5-fold effect of paxillin on AR transactivation was observed when 1 μg of paxillin expression plasmid was cotransfected with AR and the MMTV reporter (Fig. 5,A). Paxillin did not exert any effect on the basal activity of the MMTV promoter. It was not possible to assess whether paxillin effects could be saturated because inclusion of additional DNA in the transfections resulted in reduced cell viability (data not shown). The coactivator activity of paxillin was also exerted on transiently transfected GR (Fig. 5,B) but not on ERα (Fig. 5,C). This selectivity of paxillin effects is analogous to that observed with Hic-5/ARA55, which was found to act as a coactivator for GR and AR but not ERα (28). As shown in Fig. 6, paxillin is also a coactivator for AR and GR in transiently transfected PC-3 prostate cancer cells.
Functional Interactions of Paxillin with Other AR Coactivators.
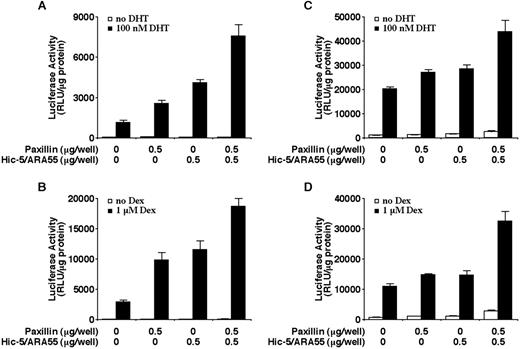
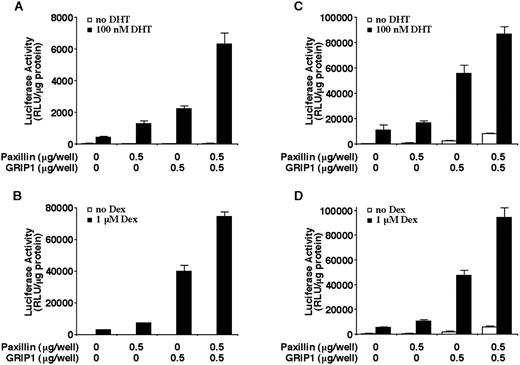
To reveal the relationship between paxillin and other nuclear receptor coactivators, we examined the effects on AR transactivation in cells cotransfected with paxillin and Hic-5/ARA55 or GRIP-1. GRIP-1 is a member of the p160 family of nuclear receptor coactivators (52) and is unrelated in structure and function to Hic-5/ARA55 and paxillin. As shown in Fig. 7, when expressed together in either CV-1 or PC-3 cells, paxillin and Hic-5/ARA55 generally exert additive effects on AR and GR transactivation. In CV-1 cells, paxillin and Hic5/ARA55 effects on AR transactivation are slightly synergistic (Fig. 7, A and B). Analogous to results obtained with Hic-5/ARA55 (28), cotransfection of paxillin with GRIP-1 also generated a synergistic effect on AR and GR transactivation in both AR and GR (Fig. 8).
Domains of Paxillin That Contribute to AR Coactivation Activity.
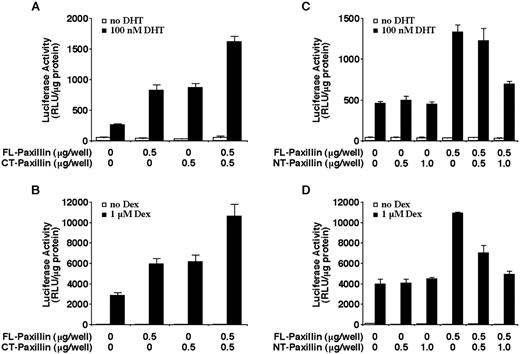
The group 3 LIM domain proteins share extensive homology within their COOH-terminal LIM domain region but possess divergent NH2-terminal domains (34). To identify the domain responsible for steroid receptor coactivator activity, paxillin NH2- and COOH-terminal domains were transfected with AR into CV-1 cells. Western blot analysis showed that both domains of paxillin were equivalently expressed in transfected cells (data not shown). As shown in Fig. 9, A and B, the COOH-terminal LIM domain of paxillin was as effective as FL paxillin in potentiating the transactivation activity of AR and GR in CV-1 cells. Cotransfection of equal amounts of FL and COOH-terminal paxillin generated additive effects on AR and GR transactivation (Fig. 9, A and B). Basal activity of the MMTV promoter (Fig. 9) was not affected by COOH-terminal paxillin. In analogous transfection experiments, the COOH-terminal region of Hic-5/ARA55 was not found to exert any effects on GR transactivation (28).
In contrast to the effect of the COOH-terminal region of paxillin, overexpression of the NH2-terminal region of paxillin did not potentiate AR transactivation in CV-1 cells. However, the NH2-terminal domain of paxillin was not neutral because its overexpression interfered with the coactivator activity of transfected FL paxillin on AR and GR (Fig. 9, C and D). This “dominant negative” effect of the NH2-terminal region of paxillin did not result from reduced expression of transfected FL paxillin (data not shown). Because AR and GR transactivation activity was not reduced by NH2-terminal paxillin beyond that observed in the absence of overexpressed paxillin (Fig. 9, C and D), this domain of paxillin may not affect the functioning of other coactivators, at least at the levels expressed under these transfection conditions.
Interaction between Paxillin and AR.
To assess whether paxillin effects on AR and GR are direct, we used a GST pull-down assay to examine paxillin interactions with these receptors. As shown in Fig. 10,A, a FL paxillin-GST fusion protein bound to AR or GR expressed in transfected CV-1 cells or the GrH2 rat hepatoma cell line, respectively. Control pull-down assays with GST alone did not result in the recovery of GR or AR, whereas the FAK protein was recovered with GST-paxillin pull-downs, as expected (Fig. 10, A and B). Importantly, the in vitro interaction between the steroid receptors (i.e., AR and GR) and paxillin was equally efficient when receptors were unliganded or hormone bound (Fig. 10, A and B). As shown in Fig. 10, C and D, the interaction between the steroid receptors (i.e., AR and GR) and paxillin occurred through its COOH-terminal LIM domain. This result is consistent with previous domain mapping of Hic-5/ARA55, which delineated its COOH-terminal LIM domain region as a steroid receptor-interaction domain (28).
DISCUSSION
LIM proteins are classified into three broad groups based largely on their localization, known partners, and apparent biological function (34). Group 1 LIM proteins localize primarily within the nucleus and affect transcription directly through their associated homeodomains or by acting as transcriptional cofactors. LIM proteins in this group are important regulators of development in various tissues (53, 54). The group 3 LIM proteins have been characterized by their association with cytoskeletal elements and predominant cytoplasmic localization (34). However, there is increasing evidence for nuclear localization of these proteins, where they might exert direct or indirect effects on transcription. In this report, we establish that paxillin, the founding member of this family, can be localized within the nucleus and in particular possesses nuclear matrix binding activity. In addition, we also show that paxillin can act as a coactivator for AR and GR, but not ERα. These properties of paxillin are analogous to those revealed previously for the group 3 LIM domain protein, Hic-5/ARA55 (27, 28). Although other members of the group 3 LIM domain family have not been tested for their effects on steroid hormone receptors, Trip6, a member of this family isolated as a thyroid hormone-interacting protein (40), functions as a relA coactivator (51). Thus, group 3 LIM domain proteins that localize predominantly within the cytoplasm, and particularly at cellular adhesion sites, have the capacity to accumulate within nuclei and exert direct effects on transcription.
Zyxin (36), LPP (37), and Ajuba (41) are members of the group 3 LIM domain protein family, the shuttling of which between sites of cell attachment and the nucleus have been assessed directly. In P19 embryonal carcinoma cells, Ajuba shuttles from sites of cell adhesion to the nucleus in response to retinoic acid-induced differentiation into an endodermal cell type (41). Although paxillin is expressed early in embryogenesis and is essential for early embryonic development (55), its localization in response to differentiation cues in vitro and in vivo has not been examined. Given the effects of paxillin and Hic-5/ARA55 on steroid receptor transactivation, the subcellular localization of these proteins may be controlled at multiple levels in developing versus fully differentiated cells. Although it is unclear whether the subcellular localization of paxillin and/or Hic-5/ARA55 can be regulated by cell adhesion or attachment in steroid hormone-responsive tissue, these dually localized coactivators could provide a direct route for extracellular cues to be transmitted to nuclear hormone receptors.
Paxillin and Hic-5/ARA55 participate in opposing cellular responses. Specifically, paxillin is more often associated with proliferative signals (32), whereas Hic5/ARA55 expression coincides with growth inhibition and cellular senescence (49). Hic-5/ARA55 and paxillin are expressed in a wide variety of vertebrate tissues, yet it is unclear how their effects on similar downstream targets bring about diverse biological responses. Overexpression of Hic-5/ARA55 in 293T cells inhibits integrin-mediated tyrosine phosphorylation of paxillin, an effect most likely mediated by competition for FAK binding (49). However, as shown in our studies, paxillin and Hic-5/ARA55 do not exhibit opposing actions on GR and AR coactivation. Rather, when expressed in limiting amounts Hic-5 and paxillin exert additive effects on steroid receptor coactivation. This observation suggests that Hic-5 and paxillin share some common mechanism in their effects on steroid receptor transactivation.
Although both Hic-5/ARA55 and paxillin use their COOH-terminal LIM domain region for interaction with AR and GR, there appears to be some distinction in their nuclear receptor coactivation. For example, although the COOH-terminal LIM domains of paxillin are as efficient as FL paxillin in potentiating GR and AR transactivation, this domain of Hic-5/ARA55 does not exhibit coactivator activity on these same receptors in transiently transfected cells (28). The NH2-terminal domain of paxillin is likewise distinguished from that of Hic-5/ARA55, given its dominant-negative effect of paxillin coactivation. The LIM domains of paxillin and Hic-5/ARA55 are highly related (30), but it is not unprecedented for highly related LIM domains to differ in their actions on transcription. For example, closely related LIM domains included within different members of the FHL family of LIMO proteins exhibit vastly different transactivation properties when tested within a single cell type (56). Extensive mutagenesis of the ACT protein member of the FHL family revealed that a precise arrangement of specific LIM domains plays a critical role in its ability to act as a transcriptional coactivator for the CREB and CREM proteins (56). This feature of the LIM domain proteins may restrict the utility of domain swaps to delineate the basis for differential effects of paxillin versus Hic-5/ARA55 COOH-terminal LIM domains on AR and GR coactivation.
The FHL2 member of the LIMO family has been found to be a very selective AR coactivator and has a restricted tissue distribution (57). These properties distinguish FHL2 from paxillin and Hic-5/ARA55, which do not share AR specificity as coactivators and are expressed in a wide range of tissues and cell types (27, 28, and this report). This highlights the versatility of LIM protein involvement in steroid receptor transactivation.
The interaction that we observed in vitro between paxillin is hormone independent. Although this appears to contrast the hormone-dependent in vitro interaction between Hic-5/ARA55 and AR (27), we in fact did not observe a hormone dependence of Hic-5/ARA55 interactions in vitro with another steroid receptor protein, GR.5 It is unclear whether the differences in hormone dependence of LIM domain protein interaction with steroid receptors reflect an artifact of extract preparation or is biologically relevant. In this regard, we note that FHL2, another recently identified LIM domain-containing AR coactivator, also did not exhibit hormone dependence in its interaction with AR in vitro (57). Analogous to Hic-5/ARA55 and paxillin, FHL2 interactions with AR involve direct contact with its LIM domain (57).
Because we did not observe an increase in basal activity of hormone-responsive promoters in transfected cells with Hic-5/ARA55 or paxillin, functional interactions of these coactivators with steroid receptors in vivo are likely to be hormone dependent. Alternatively, unliganded AR and GR may be restricted in their ability to interact with Hic-5/ARA55 and/or paxillin because of associated heat shock proteins (58). This might also explain why AR or GR targeting to focal adhesions has not been detected, although this possibility may need to be reexamined with more sophisticated cell imaging techniques. Finally, the limited amount of Hic-5/ARA55 and paxillin localized within the nucleus could provide another means of restricting interactions of steroid receptors with these coactivators. We did not observe any effects of hormone treatment on nuclear localization or Hic-5/ARA55 or paxillin in transiently transfected cells,6 but the possibility of hormone- or cell adhesion-dependent changes in Hic-5/ARA55 and paxillin subcellular localization cannot be excluded without more thorough investigation. Overexpression of paxillin did significantly enhance AR nuclear matrix binding in transfected cells, illustrating a potential role for group 3 LIM domain proteins in steroid receptor trafficking within the nucleus.
The mechanism of LIM domain protein coactivation of steroid hormone receptors remains undefined. Within all members of LIM protein groups, the LIM domain is likely to serve as a platform for protein/protein interactions (34). Although LIM proteins that contain a homeodomain, such as Lhx-3/P-LIM, bind directly to specific transcriptional regulatory sites (59), some LIMO proteins, such as LMO2, are recruited to a specific target site via their interaction with DNA-bound transcription factors such as GATA-1 and the E47-TAL1 heterodimer (60). In this context, the LIM domain facilitates assembly of a multiprotein complex that is required for optimal transcription (56). Future studies should reveal whether the group 3 LIM domain proteins Hic-5/ARA55 and paxillin serve an analogous role to stabilize steroid receptor complexes on DNA.
Recruitment of transcriptional cofactors by individual LIM domains has also been reported. For example, transcriptional activation activity of the group 1 LIM protein, Lhx3, is repressed by RLIM, a RING finger protein that interacts with a Lhx3 LIM domain (61). The specific transcriptional repression activity of RLIM is brought about by its recruitment of corepressor complexes (61). RLIM does not interact with group 3 LIM proteins (i.e., paxillin and zyxin) in vitro (60). Lhx2, a LIM protein that like Lhx3 is involved in regulation of pituitary-specific gene expression (62), also uses a partner protein to recruit a transcriptional cofactor. In contrast to RLIM, the Lhx2 LIM domain-interacting protein, MRG1, appears to function as a coactivator through its recruitment of p300/CBP (63). However, Hic-5/ARA55 and paxillin do not appear to function in an analogous mechanism to recruit other established coactivators. Although we have only obtained negative results in assays of Hic-5/ARA55 binding to nuclear coactivators of the p300 and p160 families,5 the synergistic effect of Hic-5/ARA55 and paxillin on GRIP coactivation supports the contention that these proteins use a distinct mechanism for coactivation. Recent independent results also suggest that Hic-5/ARA55 coactivation of AR is CBP independent (10). The LIMO proteins of the FHL family function as CREB/CREM coactivators in a CBP-independent manner (56). The coactivation activity of these LIM domain proteins may be related to the presence within the LIM sequence of autonomous transactivation domains (56). Hic-5/ARA55 also possesses an autonomous transactivation domain, but this is located within its NH2-terminal region and not its LIM domains (28). Although paxillin has not been tested for the presence of an autonomous transactivation domain, it differs from Hic-5/ARA55 in the location of its coactivation domain, which is localized within its LIM domain region.
Given the differences in biological functions of paxillin and Hic-5/ARA55, it may be overly simplistic to assume that their actions as coactivators for steroid hormone receptors are completely overlapping. Both proteins are expressed in androgen-responsive tissue (e.g., prostate), but it is well established that they respond in distinct ways to multiple signaling pathways. The divergent nature of their NH2-terminal domain may play a large role in differential responsiveness of these coactivators, but the potential impact of minor variations in LIM domain sequences cannot be ignored. Importantly, the alterations in paxillin function associated with the progression of prostate cancer cells to a metastatic state (43) necessitate a more thorough examination of the impact of this coactivator on AR action in prostate cancer cells with alternative cell migration and adhesion properties.
The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked advertisement in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
This work was supported by Grant R01 CA43047 and a fellowship from the University of Pittsburgh Cancer Institute (to M. K.).
The abbreviations used are: AR, androgen receptor; GR, glucocorticoid receptor; ER, estrogen receptor; LPP, LIM-containing lipoma-preferred partner; HA, hemagglutinin; MMTV, mouse mammary tumor virus; CMV, cytomegalovirus; FL, full length; FBS, fetal bovine serum; DHT, dihydrotestosterone; Dex, dexamethasone; PMSF, phenylmethylsulfonyl fluoride; GST, glutathione S-transferase; II2, indirect immunofluorescence; NMTS, nuclear matrix-targeting signal; FAK, focal adhesion kinase; LIMO, LIM only; CREB, cyclic AMP-responsive element binding protein; CREM, cyclic AMP-responsive element modulator; CBP, CREB binding protein.
J. Guerrero-Santoro and D. B. DeFranco, unpublished data.
J. Guerrero-Santoro and D. B. DeFranco, unpublished data.
J. Guerrero-Santoro, M. Kasai, and D. B. DeFranco, unpublished data.
IIF analysis of endogenous paxillin and Hic-5/ARA55 localization in CV-1 cells. CV-1 cells were fixed with paraformaldehyde and processed for IIF analysis using either an anti-paxillin antibody (A–C) or anti-Hic-5/ARA55 antibody (D–F). Before fixation, cells were either permeabilized with low-salt detergent buffer (CSK-extracted; B and E) or further digested with DNase I and then extracted with ammonium sulfate to generate a nuclear matrix fraction (C and F).
IIF analysis of endogenous paxillin and Hic-5/ARA55 localization in CV-1 cells. CV-1 cells were fixed with paraformaldehyde and processed for IIF analysis using either an anti-paxillin antibody (A–C) or anti-Hic-5/ARA55 antibody (D–F). Before fixation, cells were either permeabilized with low-salt detergent buffer (CSK-extracted; B and E) or further digested with DNase I and then extracted with ammonium sulfate to generate a nuclear matrix fraction (C and F).
Subcellular localization of paxillin and effects on AR nuclear matrix binding. A, proteins present in whole-cell lysates (Whole), CSK buffer-extracted supernatants (CSK), DNase I-digested supernatants (DNase I), or nuclear matrix pellets (NM) from CV-1, PC-3, and LNCaP cells were separated by SDS-PAGE and subjected to Western blot analysis using an anti-paxillin antibody. Blots were also stripped and reprobed with an anti-lamin B or anti-actin antibody. B, CV-1 cells transiently transfected with an AR and paxillin expression vector were incubated with 100 nm DHT for 2 h. Nuclear matrix fractions from transfected cells were subjected to Western blot analysis using an anti-AR and anti-lamin B antibody. AR in the nuclear matrix fraction was normalized using lamin B. ∗, P < 0.05, significantly different from the mean value of paxillin-nontransfected control; bars, SE.
Subcellular localization of paxillin and effects on AR nuclear matrix binding. A, proteins present in whole-cell lysates (Whole), CSK buffer-extracted supernatants (CSK), DNase I-digested supernatants (DNase I), or nuclear matrix pellets (NM) from CV-1, PC-3, and LNCaP cells were separated by SDS-PAGE and subjected to Western blot analysis using an anti-paxillin antibody. Blots were also stripped and reprobed with an anti-lamin B or anti-actin antibody. B, CV-1 cells transiently transfected with an AR and paxillin expression vector were incubated with 100 nm DHT for 2 h. Nuclear matrix fractions from transfected cells were subjected to Western blot analysis using an anti-AR and anti-lamin B antibody. AR in the nuclear matrix fraction was normalized using lamin B. ∗, P < 0.05, significantly different from the mean value of paxillin-nontransfected control; bars, SE.
Subcellular localization of FL COOH-terminal truncated paxillin in CV-1 cells. CV-1 cells were transiently transfected with expression plasmids for either FL HA-tagged paxillin (Lanes 1–3) or a deletion derivative of paxillin containing its COOH-terminal (CT) LIM domains (Lanes 4–6) and subjected to subcellular fractionation. The CSK buffer-extracted fraction (Lanes 1 and 4), DNase I-digested fraction (Lanes 2 and 5), and the nuclear matrix fraction (Lanes 3 and 6) prepared from transfected cells were subjected to Western blot analysis with anti-HA antibody and reprobed with anti-lamin B antibody. ∗, nonspecific band that appeared upon anti-HA antibody staining of blot.
Subcellular localization of FL COOH-terminal truncated paxillin in CV-1 cells. CV-1 cells were transiently transfected with expression plasmids for either FL HA-tagged paxillin (Lanes 1–3) or a deletion derivative of paxillin containing its COOH-terminal (CT) LIM domains (Lanes 4–6) and subjected to subcellular fractionation. The CSK buffer-extracted fraction (Lanes 1 and 4), DNase I-digested fraction (Lanes 2 and 5), and the nuclear matrix fraction (Lanes 3 and 6) prepared from transfected cells were subjected to Western blot analysis with anti-HA antibody and reprobed with anti-lamin B antibody. ∗, nonspecific band that appeared upon anti-HA antibody staining of blot.
Presence of paxillin in nuclear matrix fractions of human prostate tissue. Nuclear matrix fractions derived from normal prostates, prostate cancer (PCa), benign prostate hypertrophy (BPH), or LNCaP cells were subjected to Western blot analysis using an anti-paxillin antibody (A) or lamin B antibody (not shown). The ratio of paxillin/lamin B density on individual Western blots is shown (B).
Presence of paxillin in nuclear matrix fractions of human prostate tissue. Nuclear matrix fractions derived from normal prostates, prostate cancer (PCa), benign prostate hypertrophy (BPH), or LNCaP cells were subjected to Western blot analysis using an anti-paxillin antibody (A) or lamin B antibody (not shown). The ratio of paxillin/lamin B density on individual Western blots is shown (B).
Paxillin enhances AR and GR but not ER transactivation in CV-1 cells. CV-1 cells were transiently transfected with various amounts of a paxillin expression vector along with the appropriate luciferase reporter plasmid and AR (A), GR (B), or ER (C) expression vectors. After replacement of transfection medium with DMEM containing 5% charcoal-stripped FBS, the cells were incubated in the presence or absence of 10−7 m DHT (A), 10−6 m Dex (B), or 10−8 m 17β-estadiol (E2; C) for 24 h. Each point is the mean (bars, SD; n = 4) normalized to total protein levels in each sample.
Paxillin enhances AR and GR but not ER transactivation in CV-1 cells. CV-1 cells were transiently transfected with various amounts of a paxillin expression vector along with the appropriate luciferase reporter plasmid and AR (A), GR (B), or ER (C) expression vectors. After replacement of transfection medium with DMEM containing 5% charcoal-stripped FBS, the cells were incubated in the presence or absence of 10−7 m DHT (A), 10−6 m Dex (B), or 10−8 m 17β-estadiol (E2; C) for 24 h. Each point is the mean (bars, SD; n = 4) normalized to total protein levels in each sample.
Paxillin enhances AR and GR transactivation in the PC-3 prostate cancer cell line. PC-3 cells were transiently transfected with indicated amounts of a paxillin expression vector along with a MMTV promoter-linked luciferase reporter plasmid and an AR (A) expression vector or empty expression plasmid (B). A GR-expressing vector was not necessary for the GR transactivation assay because PC-3 cells express functional GR. After replacement of transfection medium with DMEM containing 5% charcoal-stripped FBS, the cells were incubated in the presence or absence of 10−7 m DHT (A) or 10−6 m Dex (B) for 24 h. The histograms show the means (bars, SD; n = 4) of relative luciferase activity normalized to total protein levels in each sample.
Paxillin enhances AR and GR transactivation in the PC-3 prostate cancer cell line. PC-3 cells were transiently transfected with indicated amounts of a paxillin expression vector along with a MMTV promoter-linked luciferase reporter plasmid and an AR (A) expression vector or empty expression plasmid (B). A GR-expressing vector was not necessary for the GR transactivation assay because PC-3 cells express functional GR. After replacement of transfection medium with DMEM containing 5% charcoal-stripped FBS, the cells were incubated in the presence or absence of 10−7 m DHT (A) or 10−6 m Dex (B) for 24 h. The histograms show the means (bars, SD; n = 4) of relative luciferase activity normalized to total protein levels in each sample.
Enhancement of hormone-induced AR or GR transactivation by paxillin and Hic-5/ARA55. CV-1 cells (A and B) or PC-3 cells (C and D) were transiently transfected with the indicated amounts of paxillin or Hic-5/ARA55 expression vectors along with a MMTV promoter-linked luciferase reporter plasmid and an AR (A and C) expression plasmid, GR expression plasmid (B), or empty expression plasmid (D). Cells were incubated in the presence or absence of 10−7 m DHT (A and C) or 10−6 m Dex (B and D) for 24 h. The histograms show the means (bars, SD; n = 4) of relative luciferase activity normalized to total protein levels in each sample.
Enhancement of hormone-induced AR or GR transactivation by paxillin and Hic-5/ARA55. CV-1 cells (A and B) or PC-3 cells (C and D) were transiently transfected with the indicated amounts of paxillin or Hic-5/ARA55 expression vectors along with a MMTV promoter-linked luciferase reporter plasmid and an AR (A and C) expression plasmid, GR expression plasmid (B), or empty expression plasmid (D). Cells were incubated in the presence or absence of 10−7 m DHT (A and C) or 10−6 m Dex (B and D) for 24 h. The histograms show the means (bars, SD; n = 4) of relative luciferase activity normalized to total protein levels in each sample.
Enhancement of hormone-induced AR or GR transactivation by paxillin and GRIP1. CV-1 cells (A and B) or PC-3 cells (C and D) were transiently transfected with the indicated amounts of paxillin or GRIP1 expression plasmids along with a MMTV promoter-linked luciferase reporter plasmid and an AR (A and C) expression plasmid, GR expression plasmid (B), or empty expression plasmid (D). Cells were incubated in the presence or absence of 10−7 m DHT (A and C) or 10−6 m Dex (B and D) for 24 h. The histograms show the means (bars, SD; n = 4) of relative luciferase activity normalized to total protein levels in each sample.
Enhancement of hormone-induced AR or GR transactivation by paxillin and GRIP1. CV-1 cells (A and B) or PC-3 cells (C and D) were transiently transfected with the indicated amounts of paxillin or GRIP1 expression plasmids along with a MMTV promoter-linked luciferase reporter plasmid and an AR (A and C) expression plasmid, GR expression plasmid (B), or empty expression plasmid (D). Cells were incubated in the presence or absence of 10−7 m DHT (A and C) or 10−6 m Dex (B and D) for 24 h. The histograms show the means (bars, SD; n = 4) of relative luciferase activity normalized to total protein levels in each sample.
Effect of paxillin COOH terminus or NH2 terminus on AR or GR transactivation. CV-1 cells were transiently transfected with the indicated amounts of FL paxillin, NH2-terminal (NT) paxillin, or COOH-terminal (CT) paxillin expression plasmids along with a MMTV promoter-linked luciferase reporter plasmid and AR (A and C) or GR (B and D) expression plasmids. Cells were incubated in the presence or absence of 10−7 m DHT (A and C) or 10−6 m Dex (B and D) for 24 h. The histograms show the means (bars, SD; n = 4) of relative luciferase activity normalized to total protein levels in each sample.
Effect of paxillin COOH terminus or NH2 terminus on AR or GR transactivation. CV-1 cells were transiently transfected with the indicated amounts of FL paxillin, NH2-terminal (NT) paxillin, or COOH-terminal (CT) paxillin expression plasmids along with a MMTV promoter-linked luciferase reporter plasmid and AR (A and C) or GR (B and D) expression plasmids. Cells were incubated in the presence or absence of 10−7 m DHT (A and C) or 10−6 m Dex (B and D) for 24 h. The histograms show the means (bars, SD; n = 4) of relative luciferase activity normalized to total protein levels in each sample.
Hormone-independent interaction of paxillin with steroid receptors in vitro. Cell lysates of CV-1 cells transfected with human AR or the GRH2 hepatoma cell line were prepared after either a 2-h hormone treatment or nontreatment and incubated with a glutathione-Sepharose-bound GST-FL-paxillin (A–D), GST-NT-paxillin (C and D), or GST-CT-paxillin (C and D) fusion protein or GST protein for 3 h. Proteins eluted from glutathione-Sepharose were subjected to Western blot analysis with anti-AR antibody (A and C) or anti-GR antibody (B and D), followed by reprobing with anti-FAK antibody (A and B).
Hormone-independent interaction of paxillin with steroid receptors in vitro. Cell lysates of CV-1 cells transfected with human AR or the GRH2 hepatoma cell line were prepared after either a 2-h hormone treatment or nontreatment and incubated with a glutathione-Sepharose-bound GST-FL-paxillin (A–D), GST-NT-paxillin (C and D), or GST-CT-paxillin (C and D) fusion protein or GST protein for 3 h. Proteins eluted from glutathione-Sepharose were subjected to Western blot analysis with anti-AR antibody (A and C) or anti-GR antibody (B and D), followed by reprobing with anti-FAK antibody (A and B).
Acknowledgments
We thank Drs. M. Nichols, D. Robins, M. Stallcup, K. Tachibana, S. Thomas, and K. Yamamoto for kind gifts of plasmids or antibodies. We also thank Dr. M. Stallcup for critical reading of the manuscript.